Science
Scientists Decode 1918 Influenza Virus Genome, Reveal Key Adaptations
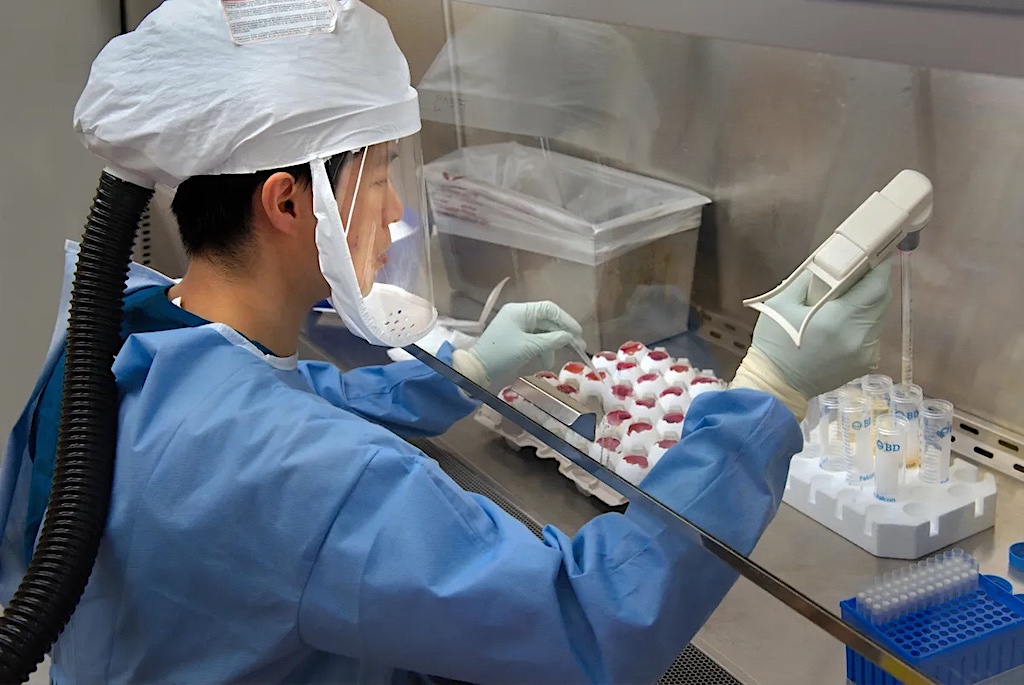
Researchers in Switzerland have made a significant breakthrough by decoding the genome of the 1918 influenza virus from a preserved patient sample in Zurich. This discovery reveals that the virus had already adapted to humans at the onset of the pandemic, carrying mutations that enhanced both its infectiousness and resistance to the immune response.
The study, conducted by the University of Zurich, utilized a novel method to recover fragile RNA from preserved tissue, providing valuable insights into the evolution of influenza viruses. This research not only sheds light on one of history’s deadliest outbreaks but also equips scientists with critical knowledge for addressing future pandemics using more informed strategies.
The Influenza Pandemic of 1918
The 1918–1920 flu pandemic, often referred to as the Great Influenza epidemic, resulted in an unprecedented loss of life, infecting nearly a third of the global population—approximately 500 million people. Death toll estimates range from 17 million to as high as 100 million, marking it as the deadliest pandemic in history. The virus responsible, identified as the H1N1 subtype of influenza A, is a negative-sense RNA virus characterized by a segmented genome. Its ability to mutate and undergo genetic reassortment has allowed it to evade host immunity and occasionally cross species barriers.
New Insights from the Swiss Genome
By comparing the Swiss strain’s genome to previously published influenza virus genomes from Germany and North America, researchers discovered that the Swiss variant possessed three critical adaptations to human hosts. Two mutations increased its resistance to an important antiviral component in the human immune system, which typically acts as a barrier against the transmission of avian-like flu viruses from animals to humans. The third mutation involved a protein in the virus’s membrane that enhanced its capacity to bind to receptors in human cells, thereby increasing its resilience and infectivity.
According to lead researcher Verena Schünemann, “This is the first time we’ve had access to an influenza genome from the 1918-1920 pandemic in Switzerland. It opens up new insights into the dynamics of how the virus adapted in Europe at the start of the pandemic.”
Implications for Future Viral Epidemics
As new viral epidemics continue to challenge public health systems worldwide, understanding how viruses evolve is essential for developing effective countermeasures. The methodology established in this research could pave the way for reconstructing additional genomes of ancient RNA viruses and verifying the authenticity of recovered RNA fragments.
Schünemann emphasized the importance of this research, stating, “A better understanding of the dynamics of how viruses adapt to humans during a pandemic over a long period of time enables us to develop models for future pandemics. Thanks to our interdisciplinary approach that combines historico-epidemiological and genetic transmission patterns, we can establish an evidence-based foundation for calculations.”
The findings of this study have been published in the journal BMC Biology, titled “An ancient influenza genome from Switzerland allows deeper insights into host adaptation during the 1918 flu pandemic in Europe.” This research not only contributes to the historical understanding of the 1918 pandemic but also serves as a vital resource for future pandemic preparedness efforts.
-

 World3 months ago
World3 months agoScientists Unearth Ancient Antarctic Ice to Unlock Climate Secrets
-

 Entertainment3 months ago
Entertainment3 months agoTrump and McCormick to Announce $70 Billion Energy Investments
-

 Lifestyle3 months ago
Lifestyle3 months agoTransLink Launches Food Truck Program to Boost Revenue in Vancouver
-

 Science3 months ago
Science3 months agoFour Astronauts Return to Earth After International Space Station Mission
-

 Technology2 months ago
Technology2 months agoApple Notes Enhances Functionality with Markdown Support in macOS 26
-

 Top Stories1 week ago
Top Stories1 week agoUrgent Update: Fatal Crash on Highway 99 Claims Life of Pitt Meadows Man
-

 Sports3 months ago
Sports3 months agoSearch Underway for Missing Hunter Amid Hokkaido Bear Emergency
-

 Politics2 months ago
Politics2 months agoUkrainian Tennis Star Elina Svitolina Faces Death Threats Online
-

 Technology3 months ago
Technology3 months agoFrosthaven Launches Early Access on July 31, 2025
-

 Politics3 months ago
Politics3 months agoCarney Engages First Nations Leaders at Development Law Summit
-

 Entertainment3 months ago
Entertainment3 months agoCalgary Theatre Troupe Revives Magic at Winnipeg Fringe Festival
-

 Politics1 week ago
Politics1 week agoShutdown Reflects Democratic Struggles Amid Economic Concerns